Running a likelihood profile on a FIMS model
likelihood_profile.Rmd
library(FIMSdiags)
library(FIMS)
packageVersion("FIMS")
#> [1] '0.7.1'
clear()Set up FIMS model
Following the minimal FIMS demo vignette, we will set up a FIMS model and run it. We need to run it initially to get the MLE parameter estimates. For this example we are using the dataset included in the FIMS package and creating a set of default parameters.
# Load sample data
data("data1")
# Prepare data for FIMS model
data_4_model <- FIMSFrame(data1)
# Create parameters
parameters <- data_4_model |>
create_default_configurations() |>
create_default_parameters(data = data_4_model)
# Run the model with optimization
base_model <- parameters |>
initialize_fims(data = data_4_model) |>
fit_fims(optimize = TRUE)
#> ✔ Starting optimization ...
#> ℹ Restarting optimizer 3 times to improve gradient.
#> ℹ Maximum gradient went from 0.02469 to 0.00037 after 3 steps.
#> ✔ Finished optimization
#> ✔ Finished sdreport
#> ℹ FIMS model version: 0.7.1
#> ℹ Total run time was 10.18995 seconds
#> ℹ Number of parameters: fixed_effects=77, random_effects=0, and total=77
#> ℹ Maximum gradient= 0.00037
#> ℹ Negative log likelihood (NLL):
#> • Marginal NLL= 3240.1261
#> • Total NLL= 3240.1261
#> ℹ Terminal SB= 1803.27712
# Clear memory post-run
clear()Run likelihood profile
Once we have a model that is fit, we can run a likelihood profile
analysis to see the impact of each data type on a given parameter. In
this example, we will test the impact on R0 but any value can
be tested by changing the parameter_name argument.
Note: The parameter name must match what it is called in the
label column of the parameters tibble. An
optional input is the module_name the parameter is
associated with. However, as long as there is only one parameter with
that name, module_name can be left NULL. The
run_fims_likelihood function takes a fitted FIMS model,
finds the MLE estimate for the parameter being profiled over (in this
case R0), and profiles over a vector of values based on the
min, max, and length the user
inputs. In the example below, it will profile over 5 values that range
from the MLE value - 1 to the MLE value + 1.
If you are running many models at once, it can be computationally more
efficient to use multiple cores. To do this, change the
n_cores argument to as many as you want/can use. NOTE:
If left empty, the default for n_cores is one less than the
number of cores on your machine so this could unexpectedly use up a lot
of your computing power.
like_fit <- run_fims_likelihood(
model = base_model,
parameters = parameters,
parameter_name = "log_rzero",
data = data1,
n_cores = 1,
min = -1,
max = 1,
length = 5
)Visualize results
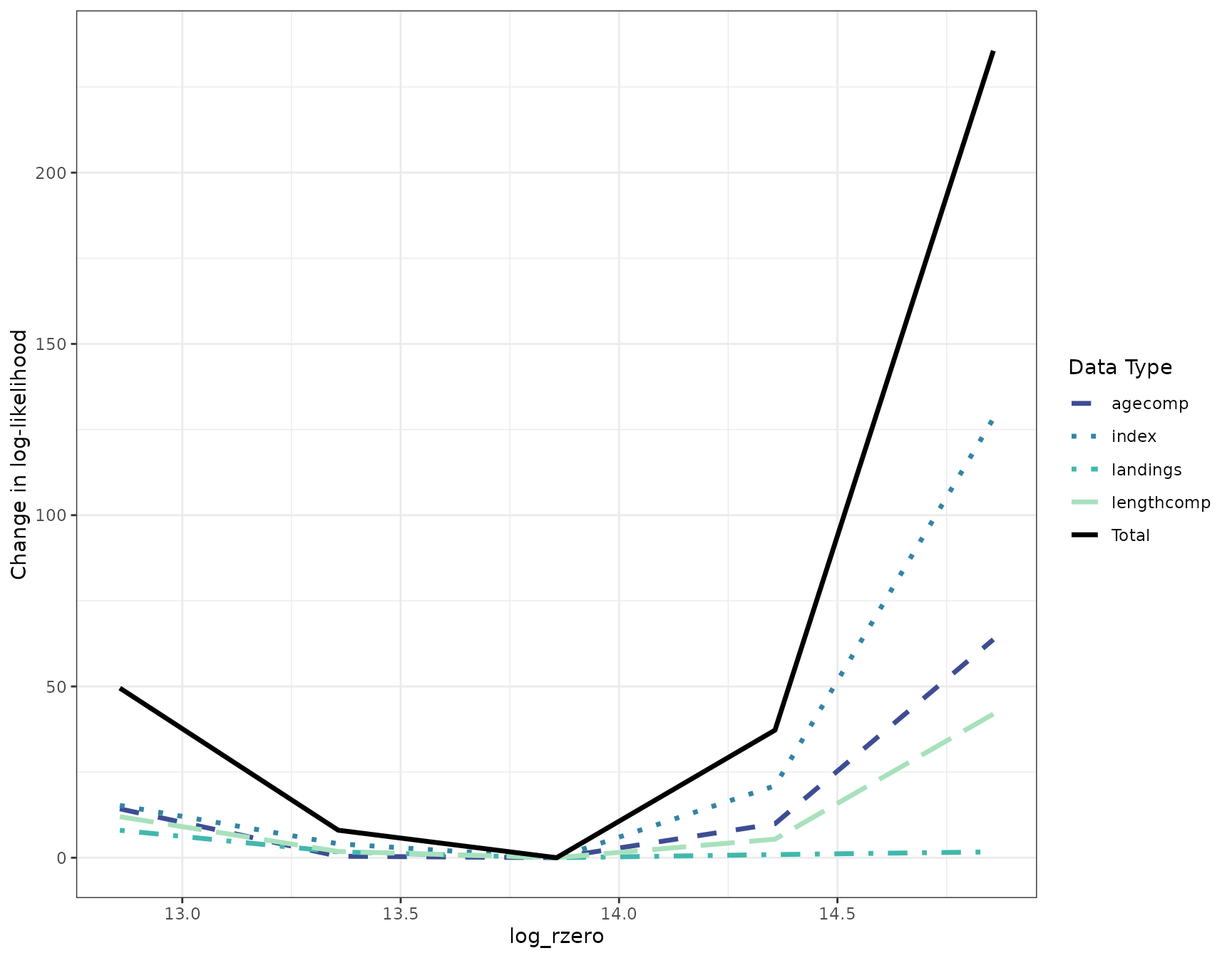
To visualize the profile, use the function
plot_likelihood. The plot shows the change in total
likelihood over the R0 values profiled in the solid black
line, and the change in likelihood for each data type in the color lines
below it. This plot indicates that the R0 value estimated by
the base model (13.9) is the value that leads to the lowest likelihood
and all of the data types support this (i.e. the data do not
conflict).
plot_likelihood(like_fit)
#> Scale for colour is already present.
#> Adding another scale for colour, which will replace the existing scale.
#> Saving 9 x 7 in image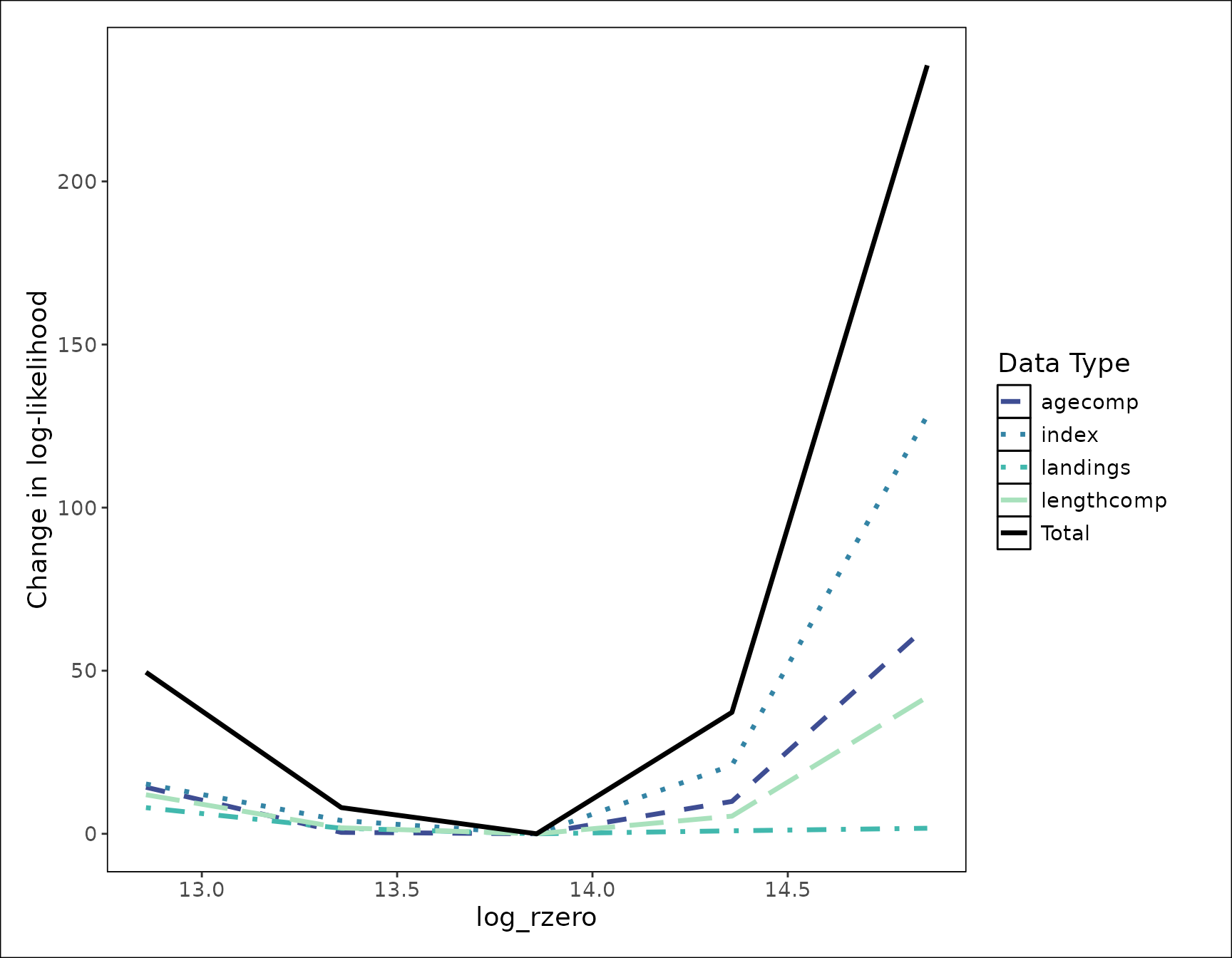
This function creates a ggplot object that can be
customized and added onto as a normal ggplot can. The
default theme is stockplotr::theme_noaa, however to change
the theme or colors used, you can simply add them after the main plot
function.
plot_likelihood(like_fit) +
ggplot2::theme_bw()
#> Scale for colour is already present.
#> Adding another scale for colour, which will replace the existing scale.
#> Saving 9 x 7 in image