Running a retrospective analysis on a FIMS model
retrospective.Rmd
library(FIMSdiags)
library(FIMS)
packageVersion("FIMS")
#> [1] '0.7.1'
clear()Set up FIMS model
Following the minimal FIMS demo vignette, we will set up a FIMS model and run it. This model will contain the all years of data and will be our reference model (see section below for more information about reference models). For this example we are using the dataset included in the FIMS package and creating a set of default parameters.
# Load sample data
data("data1")
# Prepare data for FIMS model
data_4_model <- FIMSFrame(data1)
# Create parameters
parameters <- data_4_model |>
create_default_configurations() |>
create_default_parameters(data = data_4_model)
# Run the model with optimization
base_model <- parameters |>
initialize_fims(data = data_4_model) |>
fit_fims(optimize = TRUE)
#> ✔ Starting optimization ...
#> ℹ Restarting optimizer 3 times to improve gradient.
#> ℹ Maximum gradient went from 0.02469 to 0.00037 after 3 steps.
#> ✔ Finished optimization
#> ✔ Finished sdreport
#> ℹ FIMS model version: 0.7.1
#> ℹ Total run time was 10.11825 seconds
#> ℹ Number of parameters: fixed_effects=77, random_effects=0, and total=77
#> ℹ Maximum gradient= 0.00037
#> ℹ Negative log likelihood (NLL):
#> • Marginal NLL= 3240.1261
#> • Total NLL= 3240.1261
#> ℹ Terminal SB= 1803.27712
# Clear memory post-run
clear()Retrospective analysis
A retrospective analysis is a common model diagnostic used to assess how stable model estimates such as spawning biomass and fishing mortality are and if there is a consistent pattern in the estimates as years of data are removed. The process involves removing n years of data from the end of the time series and refitting the model. This is an iterative process where the user typically runs 5-10 models, with each run (often called “peels”) progressively removing another year of data. Derived quantities such as spawning biomass, fishing mortality (F), and recruitment over the years of included data are compared to the model run with the full dataset (reference model). The Mohn’s rho statistic can be used to compare the relative difference between the reference model and each peel. Mohn’s rho statistic is calculated as:
where is the number of retrospective peels, is the estimated value (e.g. spawning biomass, fishing mortality, etc.) from peel at its terminal year , and is the value from the reference model, , at that same year .
Some things to note about the run_fims_retrospective()
function, you can provide data that is already a FIMSFrame
object or you can give it one that is not. In the example below, we are
using the FIMSFrame data object (data_4_model)
created above. Additionally, it expects a vector of positive values of
years_to_remove from the base model, starting from 0. If
you are running many peels, it may be more efficient to use multiple
cores, which can be specified by the n_cores argument.
NOTE: If left empty, the default for
n_coresis one less than the number of cores on your machine so this could unexpectedly use up a lot of your computing power.
retro_fit <- run_fims_retrospective(years_to_remove = 0:2,
data_4_model, parameters,
n_cores = 1)
#> ℹ ...Running sequentially on a single core
#> ℹ running model with 0 years of data removed
#> ✔ Starting optimization ...
#> ℹ Restarting optimizer 3 times to improve gradient.
#> ℹ Maximum gradient went from 0.02469 to 0.00037 after 3 steps.
#> ✔ Finished optimization
#> ✔ Finished sdreport
#> ℹ FIMS model version: 0.7.1
#> ℹ Total run time was 9.76148 seconds
#> ℹ Number of parameters: fixed_effects=77, random_effects=0, and total=77
#> ℹ Maximum gradient= 0.00037
#> ℹ Negative log likelihood (NLL):
#> • Marginal NLL= 3240.1261
#> • Total NLL= 3240.1261
#> ℹ Terminal SB= 1803.27712
#> ℹ running model with 1 years of data removed
#>
#> ✔ Starting optimization ...
#> ℹ Restarting optimizer 3 times to improve gradient.
#> ℹ Maximum gradient went from 0.03303 to 0.00161 after 3 steps.
#> ✔ Finished optimization
#> ✔ Finished sdreport
#> ℹ FIMS model version: 0.7.1
#> ℹ Total run time was 20.60323 seconds
#> ℹ Number of parameters: fixed_effects=77, random_effects=0, and total=77
#> ℹ Maximum gradient= 0.00161
#> ℹ Negative log likelihood (NLL):
#> • Marginal NLL= 3140.71461
#> • Total NLL= 3140.71461
#> ℹ Terminal SB= 2315.0298
#> ℹ running model with 2 years of data removed
#>
#> ✔ Starting optimization ...
#> ℹ Restarting optimizer 3 times to improve gradient.
#> ℹ Maximum gradient went from 0.01823 to 0.00303 after 3 steps.
#> ✔ Finished optimization
#> ✔ Finished sdreport
#> ℹ FIMS model version: 0.7.1
#> ℹ Total run time was 20.48512 seconds
#> ℹ Number of parameters: fixed_effects=77, random_effects=0, and total=77
#> ℹ Maximum gradient= 0.00303
#> ℹ Negative log likelihood (NLL):
#> • Marginal NLL= 3034.31988
#> • Total NLL= 3034.31988
#> ℹ Terminal SB= 3115.03058Mohn’s Rho
Once you have run a retrospective, you can then calculate the Mohn’s rho statistic for spawning biomass by:
rho_sb <- calculate_mohns_rho(retro_fit, quantity = "spawning_biomass")Visualize the results
To visualize the results of the retrospective analysis, we can use
plot_retrospective to display the quantity of interest.
Below, we want to assess the impact on spawning biomass. The function
returns a ggplot object so to customize the default figure,
you can add layers as you normally would to a ggplot. For
example, below, we specify the x and y axes labels and the legend
title.
🚧 Under development We are actively working on expanding which quantities can be visualized with the
plot_retrospectivefunction but for now, it is limited tospawning_biomass.
# plot the SSB time series from each model
fig_retrospective_ssb <- plot_retrospective(retro_fit) +
ggplot2::labs(x = "Year", y = "Spawning Biomass") +
ggplot2::guides(fill = ggplot2::guide_legend(title = "Retro Peel"),
color = ggplot2::guide_legend(title = "Retro Peel"),
linetype = ggplot2::guide_legend(title = "Retro Peel"))
#> Saving 9 x 7 in image
print(fig_retrospective_ssb)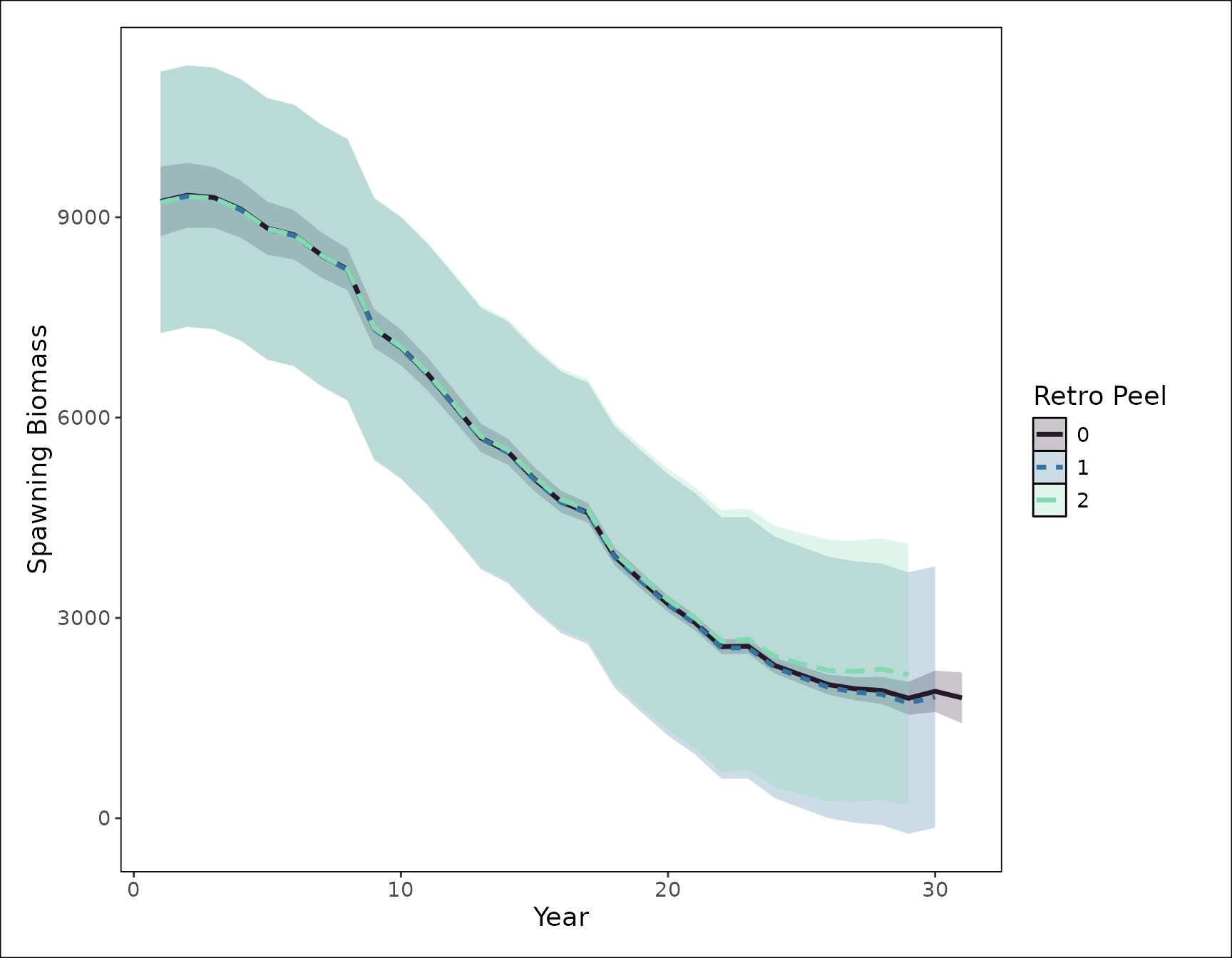
We can compare the spawning biomass estimates from the reference model and each peel for the last six years.
retro_fit[["estimates"]] |>
dplyr::filter(label == "spawning_biomass") |>
dplyr::select(label, year_i, estimated, retrospective_peel) |>
tidyr::pivot_wider(names_from = retrospective_peel, values_from = estimated) |>
dplyr::rename(
"Year" = year_i,
"Reference model" = `0`,
"Peel 1" = `1`,
"Peel 2" = `2`
) |>
tail()
#> # A tibble: 6 × 5
#> label Year `Reference model` `Peel 1` `Peel 2`
#> <chr> <int> <dbl> <dbl> <dbl>
#> 1 spawning_biomass 26 2001. 1960. 2213.
#> 2 spawning_biomass 27 1939. 1889. 2201.
#> 3 spawning_biomass 28 1914. 1854. 2232.
#> 4 spawning_biomass 29 1798. 1725. 2153.
#> 5 spawning_biomass 30 1901. 1815. 2623.
#> 6 spawning_biomass 31 1803. 2315. 3115.