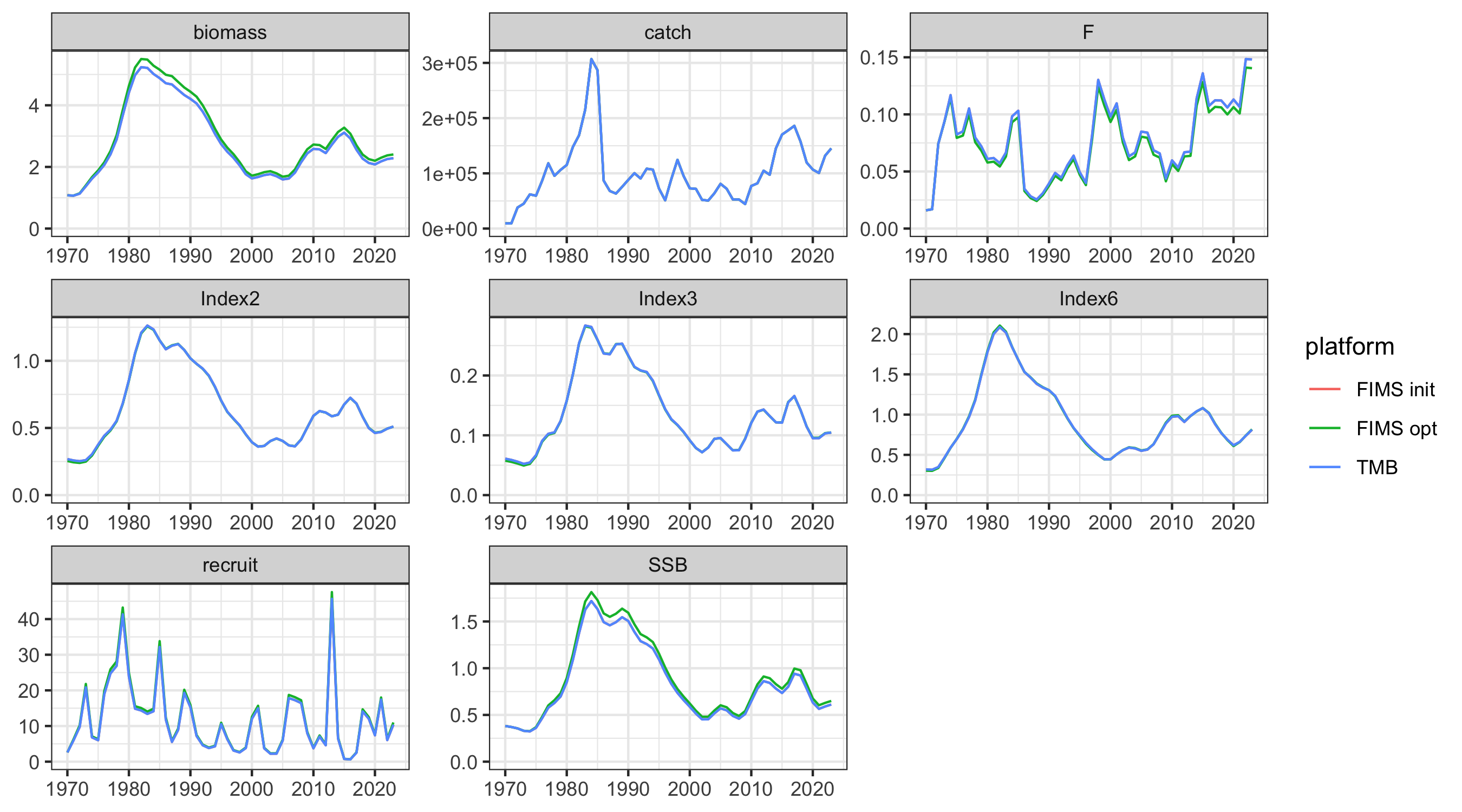
out1 <- get_long_outputs(rep1, rep0) |>
dplyr::mutate(platform = ifelse(platform == "FIMS", "FIMS init", "TMB"))
out2 <- get_long_outputs(rep2, rep0) |>
dplyr::filter(platform == "FIMS") |>
dplyr::mutate(platform = "FIMS opt")
out <- rbind(out1, out2)
g <- ggplot2::ggplot(
out,
ggplot2::aes(year, value, color = platform)
) +
ggplot2::geom_line() +
ggplot2::facet_wrap("name", scales = "free") +
ggplot2::ylim(0,NA) +
ggplot2::labs(x = NULL, y = NULL)
ggplot2::ggsave("figures/AFSC_PK_ts_comparison.png", g, width = 9, height = 5, units = "in")
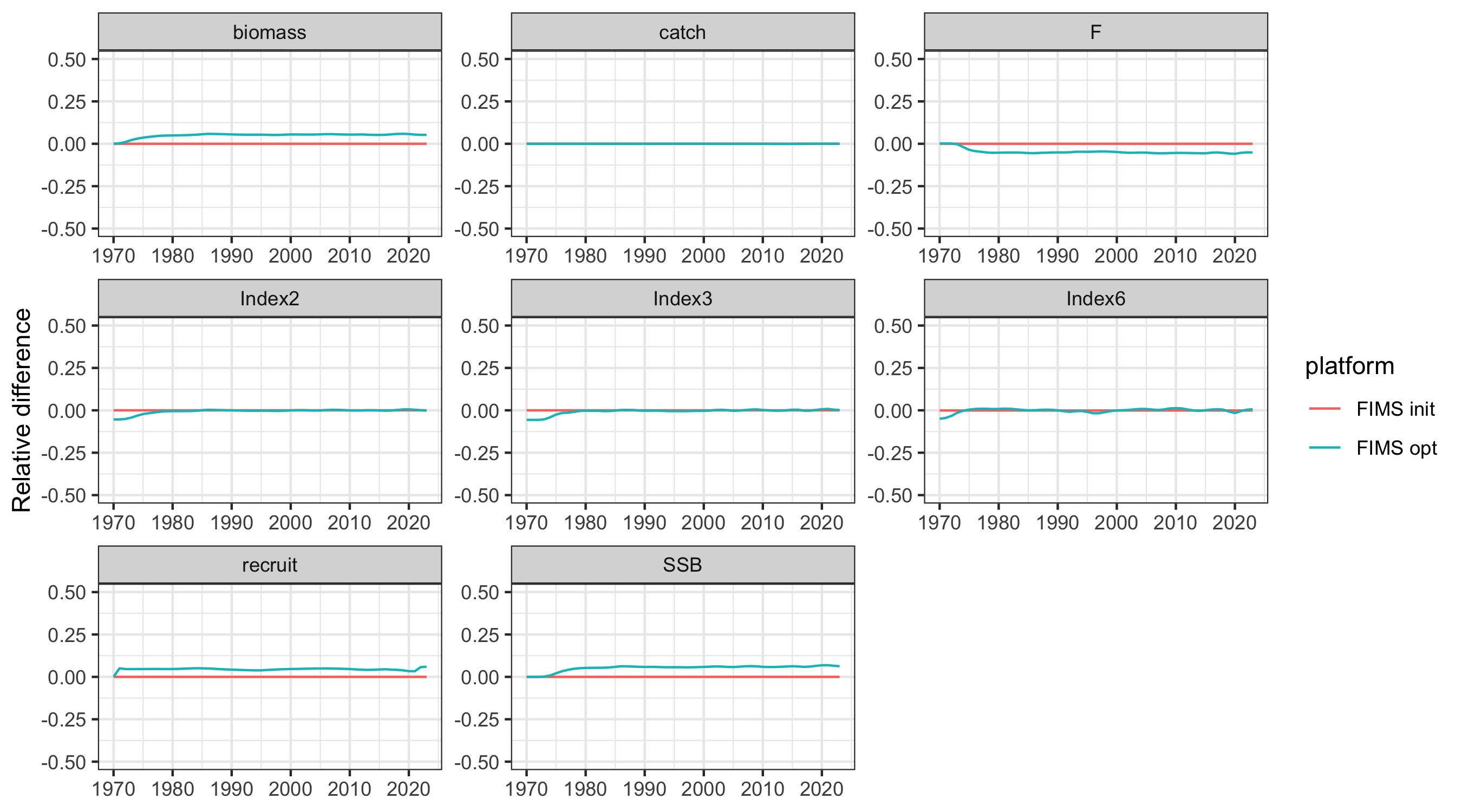
g <- ggplot2::ggplot(
dplyr::filter(out, platform != "TMB"),
ggplot2::aes(year, relerror, color = platform)
) +
ggplot2::geom_line() +
ggplot2::facet_wrap("name", scales = "free") +
ggplot2::labs(x = NULL, y = "Relative difference") +
ggplot2::coord_cartesian(ylim = c(-.5,.5))
ggplot2::ggsave("figures/AFSC_PK_ts_comparison_relerror.png", g, width = 9, height = 5, units = "in")
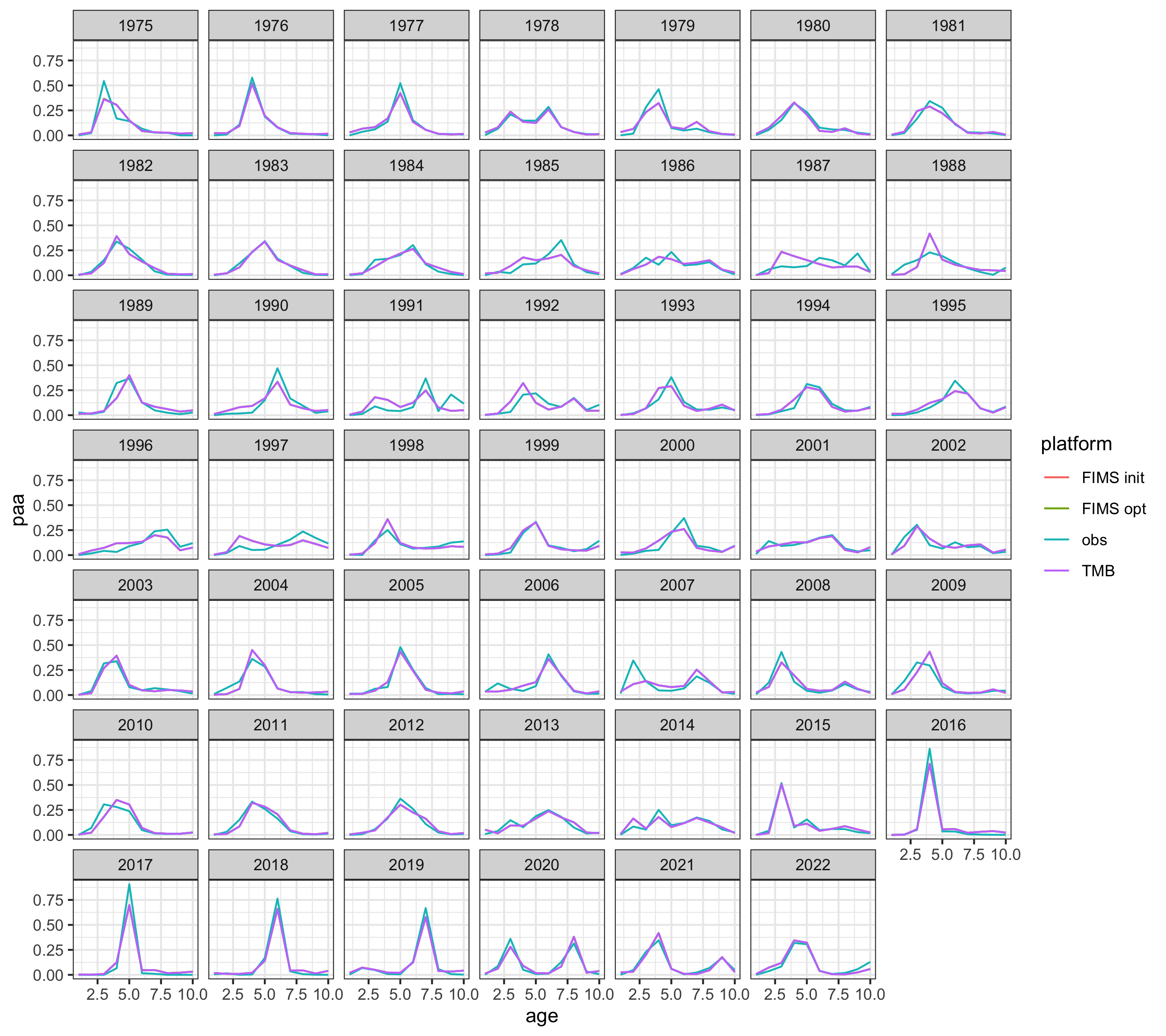
## Quick check on age comp fits
p1 <- get_acomp_fits(rep0, rep1, rep2, fleet = 1, years = pkdat0$fshyrs)
g <- ggplot2::ggplot(p1, ggplot2::aes(age, paa, color = platform)) +
ggplot2::facet_wrap("year") +
ggplot2::geom_line()
ggplot2::ggsave("figures/AFSC_PK_age_comp_fits_1.png", g, width = 9, height = 8, units = "in")
p2 <- get_acomp_fits(rep0, rep1, rep2, fleet = 2, years = pkdat0$srv_acyrs2)
g <- ggplot2::ggplot(p2, ggplot2::aes(age, paa, color = platform)) +
ggplot2::facet_wrap("year") +
ggplot2::geom_line()
ggplot2::ggsave("figures/AFSC_PK_age_comp_fits_2.png", g, width = 9, height = 8, units = "in")
## p3 <- get_acomp_fits(rep0, rep1, rep2, fleet = 3, years = pkdat0$srv_acyrs3)
## g <- ggplot2::ggplot(p3, ggplot2::aes(age, paa, color = platform)) + ggplot2::facet_wrap("year") + ggplot2::geom_line()
## p6 <- get_acomp_fits(rep0, rep1, rep2, fleet = 4, years = pkdat0$srv_acyrs6)
## g <- ggplot2::ggplot(p6, ggplot2::aes(age, paa, color = platform)) + ggplot2::facet_wrap("year") + ggplot2::geom_line()
## index fits
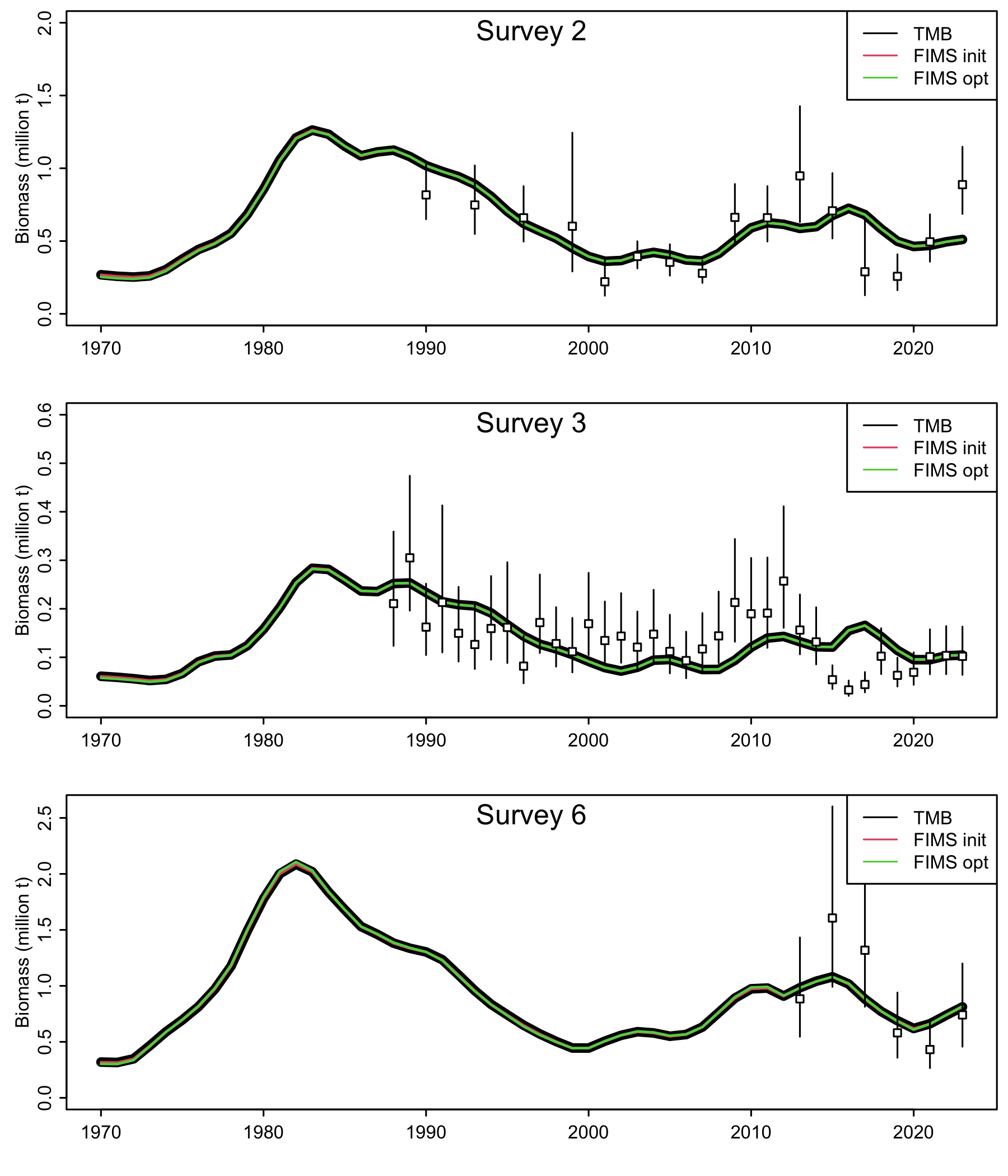
addsegs <- function(yrs, obs, CV){
getlwr <- function(obs, CV) qlnorm(p = .025, meanlog = log(obs), sdlog = sqrt(log(1+CV^2)))
getupr <- function(obs, CV) qlnorm(p = .975, meanlog = log(obs), sdlog = sqrt(log(1+CV^2)))
segments(yrs, y0 = getlwr(obs,CV), y1 = getupr(obs,CV))
points(yrs, obs, pch = 22, bg = "white")
}
png("figures/AFSC_PK_index_fits.png", res = 300, width = 6, height = 7, units = "in")
par(mfrow = c(3,1), mar = c(3,3,.5,.5), mgp = c(1.5,.5,0), tck = -0.02)
plot(years, rep0$Eindxsurv2, type = "l",
ylim = c(0,2), lwd = 5.5,
xlab = NA, ylab = "Biomass (million t)")
x1 <- out1 |>
dplyr::filter(name == "Index2" & platform == "FIMS init")
x2 <- out2 |>
dplyr::filter(name == "Index2" & platform == "FIMS opt")
lines(years,x1$value, col = 2, lwd = 1.5)
lines(years,x2$value, col = 3, lwd = 1.5)
addsegs(yrs = pkdat0$srvyrs2, obs = pkdat0$indxsurv2, CV = pkdat0$indxsurv_log_sd2)
legend("topright", legend = c("TMB", "FIMS init", "FIMS opt"), lty = 1, col = 1:3)
mtext("Survey 2", line = -1.5)
plot(years, rep0$Eindxsurv3, type = "l",
ylim = c(0,.6), lwd = 5.5,
xlab = NA, ylab = "Biomass (million t)")
x1 <- out1 |>
dplyr::filter(name == "Index3" & platform == "FIMS init")
x2 <- out2 |>
dplyr::filter(name == "Index3" & platform == "FIMS opt")
lines(years,x1$value, col = 2, lwd = 1.5)
lines(years,x2$value, col = 3, lwd = 1.5)
addsegs(yrs = pkdat0$srvyrs3, obs = pkdat0$indxsurv3, CV = pkdat0$indxsurv_log_sd3)
mtext("Survey 3", line = -1.5)
legend("topright", legend = c("TMB", "FIMS init", "FIMS opt"), lty = 1, col = 1:3)
plot(years, rep0$Eindxsurv6, type = "l",
ylim = c(0,2.6), lwd = 5.5,
xlab = NA, ylab = "Biomass (million t)")
x1 <- out1 |>
dplyr::filter(name == "Index6" & platform == "FIMS init")
x2 <- out2 |>
dplyr::filter(name == "Index6" & platform == "FIMS opt")
lines(years,x1$value, col = 2, lwd = 1.5)
lines(years,x2$value, col = 3, lwd = 1.5)
addsegs(yrs = pkdat0$srvyrs6, obs = pkdat0$indxsurv6, CV = pkdat0$indxsurv_log_sd6)
mtext("Survey 6", line = -1.5)
legend("topright", legend = c("TMB", "FIMS init", "FIMS opt"), lty = 1, col = 1:3)
dev.off()