######################################################################
# Plot results
# created by running colorspace::sequential_hcl(5, "Viridis")
cols <- c("#4B0055", "#00588B", "#009B95", "#53CC67", "#FDE333")
out.folder <- "figures"
dir.create(out.folder, showWarnings = FALSE)
plot.type <- "png"
get_selectivity_parameter <- function(fit, x) {
good <- dplyr::filter(
FIMS::get_estimates(fit),
module_name == "Selectivity",
label == x
)
out <- dplyr::pull(good, estimated)
names(out) <- paste(
dplyr::pull(good, label),
dplyr::pull(good, parameter_id),
sep = "_"
)
return(out)
}
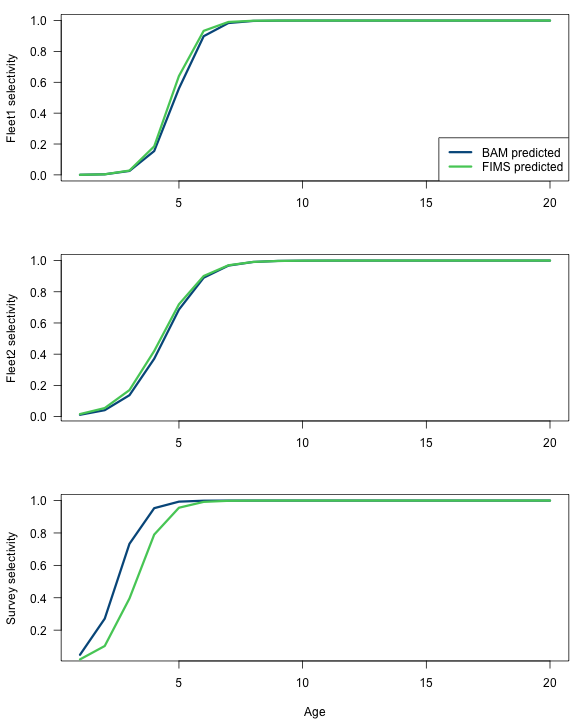
selex.bam.fleet1 <- 1 / (1 + exp(-sca$parm.cons$selpar_slope_COM2[8] * (ages - sca$parm.cons$selpar_A50_COM2[8])))
selex.fims.fleet1 <- 1 / (1 + exp(-get_selectivity_parameter(fit, "slope")[1] * (ages - get_selectivity_parameter(fit, "inflection_point")[1])))
selex.bam.fleet2 <- 1 / (1 + exp(-sca$parm.cons$selpar_slope1_REC2[8] * (ages - sca$parm.cons$selpar_A50_REC2[8])))
selex.fims.fleet2 <- 1 / (1 + exp(-get_selectivity_parameter(fit, "slope")[2] * (ages - get_selectivity_parameter(fit, "inflection_point")[2])))
selex.bam.survey <- 1 / (1 + exp(-sca$parm.cons$selpar_slope1_CVT[8] * (ages - sca$parm.cons$selpar_A501_CVT[8])))
selex.fims.survey <- 1 / (1 + exp(-get_selectivity_parameter(fit, "slope")[3] * (ages - get_selectivity_parameter(fit, "inflection_point")[3])))
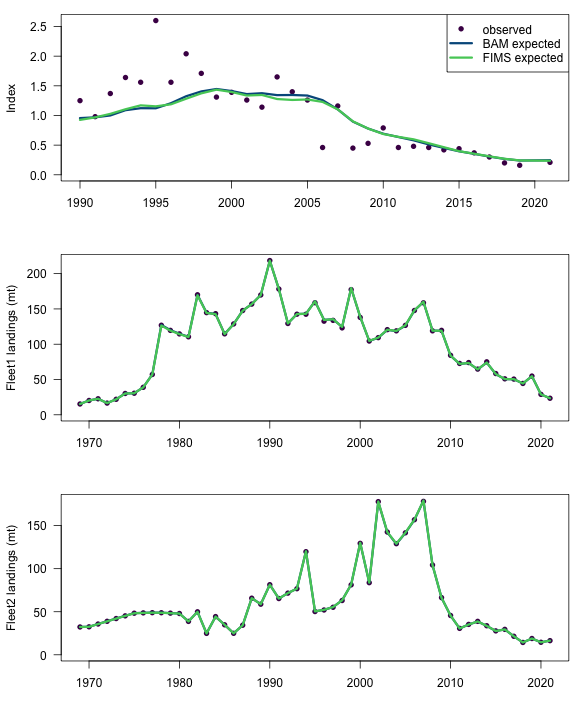
index_results_allyr <- data.frame(
yr = styr:endyr,
observed = FIMS::get_data(data_4_model) |>
dplyr::filter(type == "index", name == "survey1") |>
dplyr::pull(value),
fims.expected = report$index_expected[[3]],
bam.expected = sca$t.series$U.CVT.pr
)
index_results <- index_results_allyr |>
dplyr::filter(observed != -999.00)
fleet1_landings_results <- data.frame(
yr = styr:endyr,
observed = FIMS::get_data(data_4_model) |>
dplyr::filter(type == "landings", name == "fleet1") |>
dplyr::pull(value),
fims.expected = report$landings_expected[[1]],
bam.expected = sca$t.series$L.COM.pr
)
fleet2_landings_results <- data.frame(
yr = styr:endyr,
observed = FIMS::get_data(data_4_model) |>
dplyr::filter(type == "landings", name == "fleet2") |>
dplyr::pull(value),
fims.expected = report$landings_expected[[2]],
bam.expected = sca$t.series$L.REC.pr
)
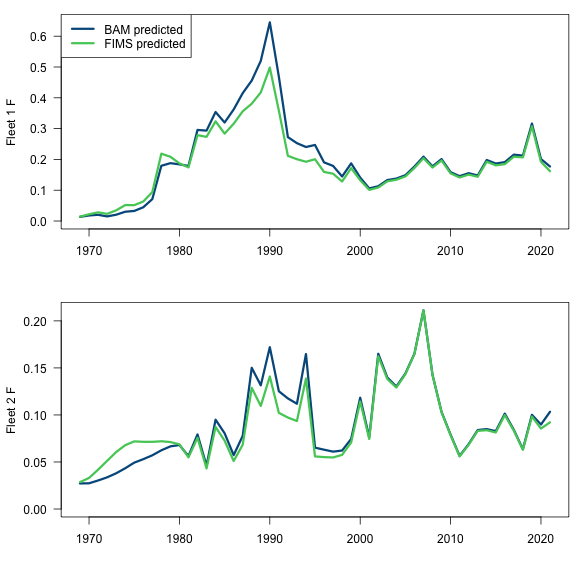
fleet1_F_results <- data.frame(
yr = styr:endyr,
fims.F.fleet1 = dplyr::filter(FIMS::get_estimates(fit), label == "log_Fmort", module_id == 1) |> dplyr::pull(estimated),
bam.F.fleet1 = sca$t.series$F.COM
)
fleet2_F_results <- data.frame(
yr = styr:endyr,
fims.F.fleet2 = dplyr::filter(FIMS::get_estimates(fit), label == "log_Fmort", module_id == 2) |> dplyr::pull(estimated),
bam.F.fleet2 = sca$t.series$F.REC
)
# Dropping the last (extra) year from FIMS output, assuming it is a projection yr (not an initialization yr)
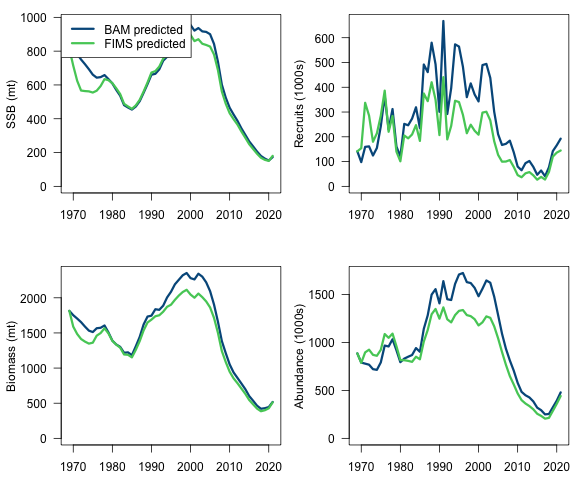
fims.naa <- matrix(report$numbers_at_age[[1]], ncol = n_ages, byrow = TRUE)
fims.naa <- fims.naa[-54, ]
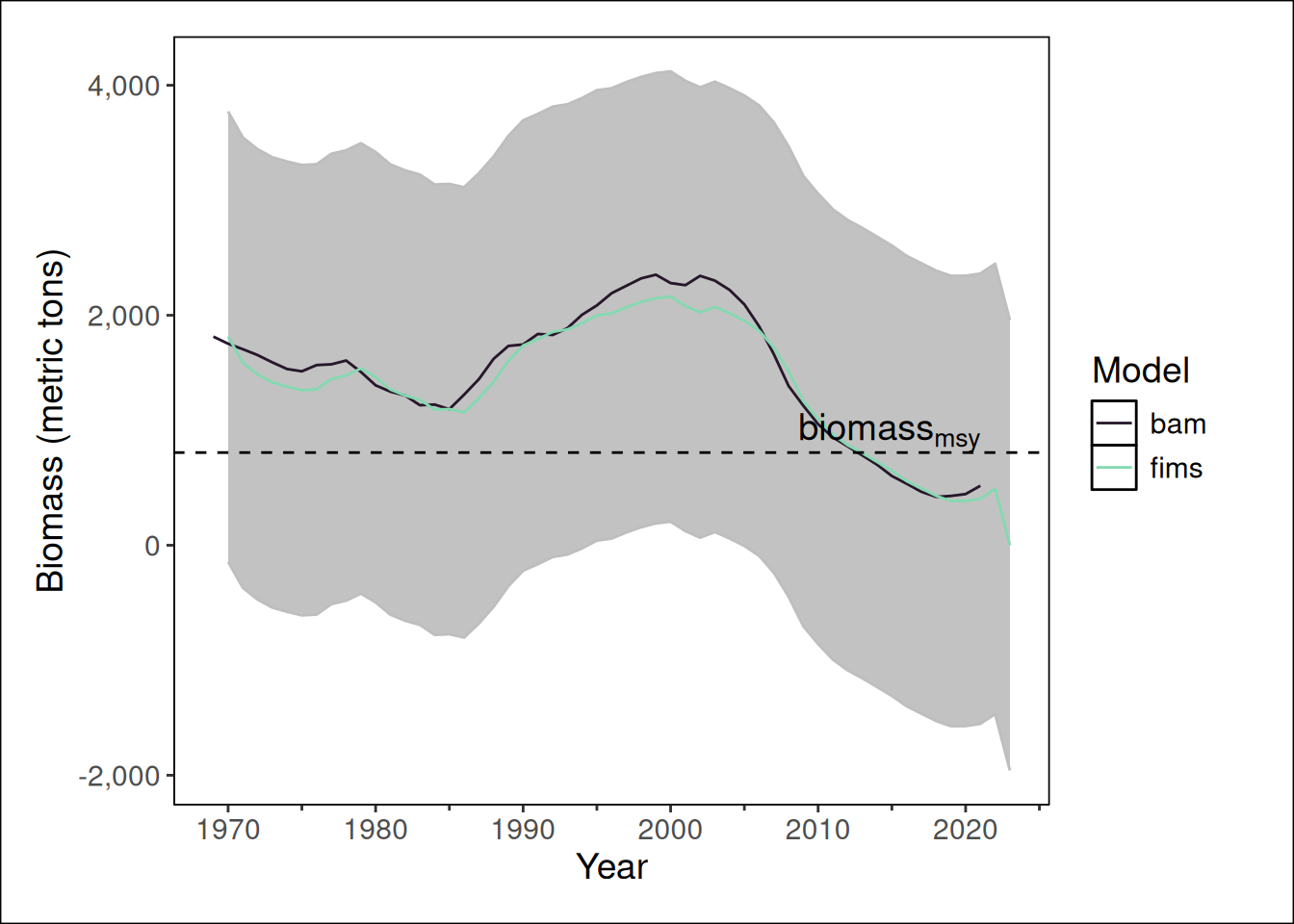
popn_results <- data.frame(
yr = styr:endyr,
fims.ssb = report$spawning_biomass[[1]][1:n_years],
fims.recruits = report$expected_recruitment[[1]][1:n_years] / 1000,
fims.biomass = report$biomass[[1]][1:n_years],
fims.abundance = rowSums(fims.naa) / 1000,
bam.ssb = sca$t.series$SSB,
bam.recruits = sca$t.series$recruits / 1000,
bam.biomass = sca$t.series$B,
bam.abundance = sca$t.series$N / 1000
)
yr.ind <- 1:n_years
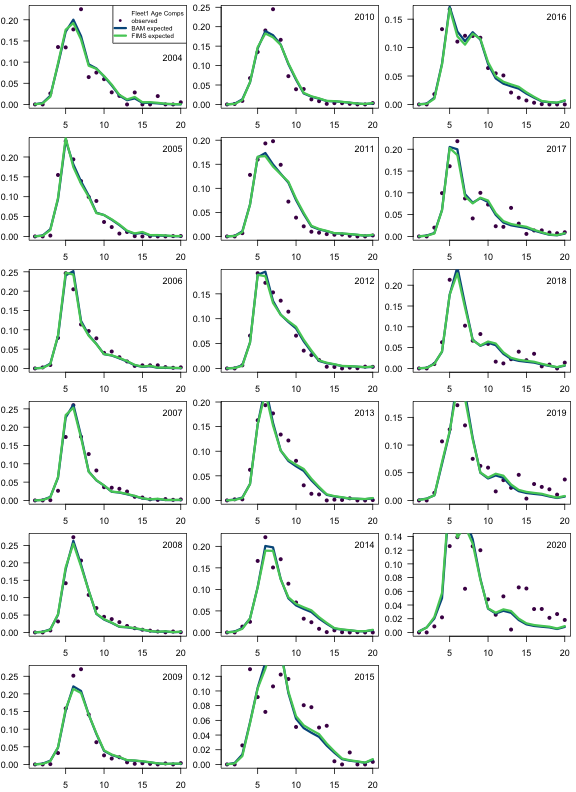
yr.fleet1.ind <- yr.ind[fleet1_ac_n >= 0]
yr.fleet1.ac <- years[yr.fleet1.ind]
fims.fleet1.ncaa <- matrix(report$landings_numbers_at_age[[1]], ncol = n_ages, byrow = TRUE)
fims.fleet1.ncaa <- fims.fleet1.ncaa[yr.fleet1.ind, ]
fims.fleet1.caa <- fims.fleet1.ncaa / rowSums(fims.fleet1.ncaa)
bam.fleet1.caa <- sca$comp.mats$acomp.COM.pr
obs.fleet1.caa <- sca$comp.mats$acomp.COM.ob
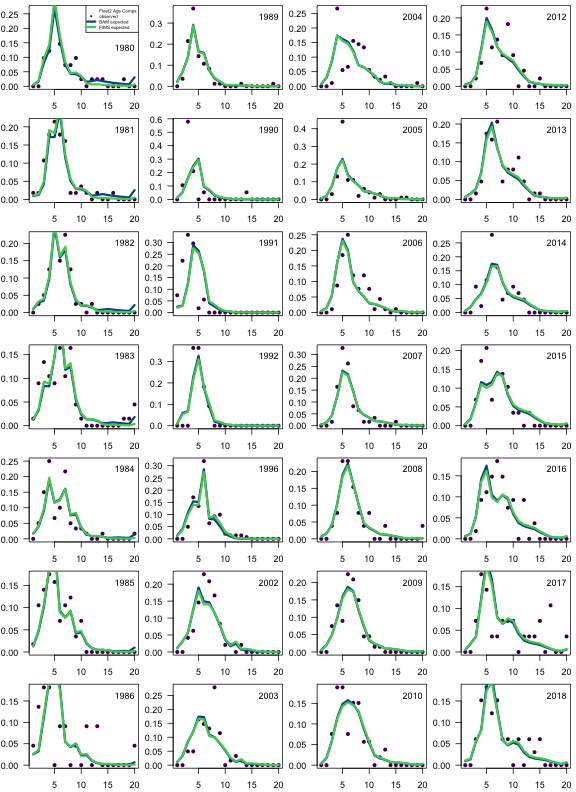
yr.fleet2.ind <- yr.ind[fleet2_ac_n >= 0]
yr.fleet2.ac <- years[yr.fleet2.ind]
fims.fleet2.ncaa <- matrix(report$landings_numbers_at_age[[2]], ncol = n_ages, byrow = TRUE)
fims.fleet2.ncaa <- fims.fleet2.ncaa[yr.fleet2.ind, ]
fims.fleet2.caa <- fims.fleet2.ncaa / rowSums(fims.fleet2.ncaa)
bam.fleet2.caa <- sca$comp.mats$acomp.REC.pr
obs.fleet2.caa <- sca$comp.mats$acomp.REC.ob
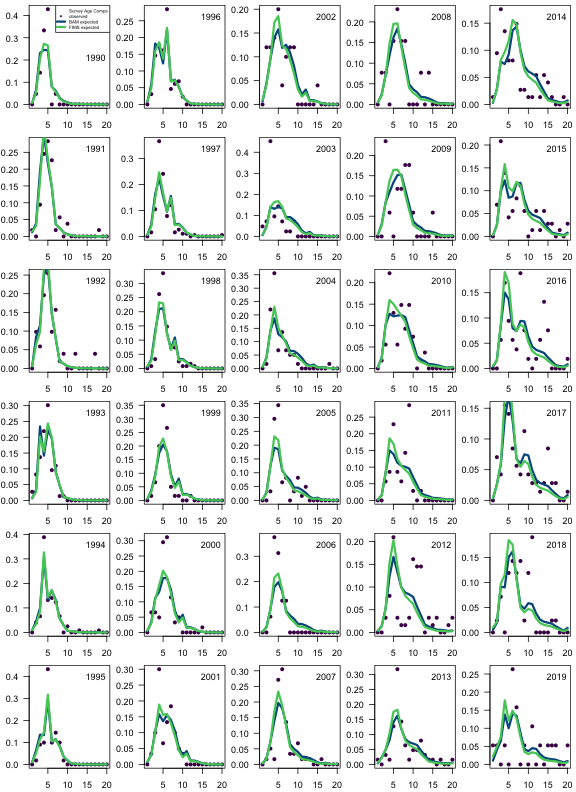
yr.survey.ind <- yr.ind[survey_ac_n >= 0]
yr.survey.ac <- years[yr.survey.ind]
fims.survey.ncaa <- matrix(report$landings_numbers_at_age[[3]], ncol = n_ages, byrow = TRUE)
fims.survey.ncaa <- fims.survey.ncaa[yr.survey.ind, ]
fims.survey.caa <- fims.survey.ncaa / rowSums(fims.survey.ncaa)
bam.survey.caa <- sca$comp.mats$acomp.CVT.pr
obs.survey.caa <- sca$comp.mats$acomp.CVT.ob
######################################################################
png(filename = paste(out.folder, "/SEFSC_scamp_tseries_fits.", plot.type, sep = ""), width = 8, height = 10, units="in", res=72)
mat <- matrix(1:3, ncol = 1)
layout(mat = mat, widths = rep.int(1, ncol(mat)), heights = rep.int(1, nrow(mat)))
par(las = 1, mar = c(4.1, 4.25, 1.0, 0.5), cex = 1)
plot(index_results$yr, index_results$observed,
ylim = c(0, max(index_results[, -1])),
pch = 16, col = cols[1], ylab = "Index", xlab = ""
)
lines(index_results$yr, index_results$bam.expected, lwd = 3, col = cols[2])
lines(index_results$yr, index_results$fims.expected, lwd = 3, col = cols[4])
legend("topright",
legend = c("observed", "BAM expected", "FIMS expected"),
pch = c(16, -1, -1), lwd = c(-1, 3, 3), col = c(cols[1], cols[2], cols[4])
)
plot(fleet1_landings_results$yr, fleet1_landings_results$observed,
ylim = c(0, max(fleet1_landings_results[, -1])),
pch = 16, col = cols[1], ylab = "Fleet1 landings (mt)", xlab = ""
)
lines(fleet1_landings_results$yr, fleet1_landings_results$bam.expected, lwd = 3, col = cols[2])
lines(fleet1_landings_results$yr, fleet1_landings_results$fims.expected, lwd = 3, col = cols[4])
plot(fleet2_landings_results$yr, fleet2_landings_results$observed,
ylim = c(0, max(fleet2_landings_results[, -1])),
pch = 16, col = cols[1], ylab = "Fleet2 landings (mt)", xlab = ""
)
lines(fleet2_landings_results$yr, fleet2_landings_results$bam.expected, lwd = 3, col = cols[2])
lines(fleet2_landings_results$yr, fleet2_landings_results$fims.expected, lwd = 3, col = cols[4])
dev.off()